Using Supercomputers and Machine Learning to Discover Defective Amino Acids that Cause Diseases
Tiny defects under high-tech observation could point to breakthroughs.

Many diseases including cancer, diabetes and digestive disorders are caused by malfunctioning ribosomes and proteins. In the human body, ribosomes provide codes for building proteins. A research team led by Narayana R. Aluru, Ph.D., M.S., from the University of Illinois at Urbana-Champaign, Department of Mechanical Science and Engineering, Beckman Institute for Advanced Science and Technology is doing research on amino acids to help locate faulty amino acids and proteins.
The miniscule defects under high-tech observation could potentially point the way to medical breakthroughs, according to experts.
“Many diseases are caused by the faulty reading of DNA in the ribosomes which leads to a faulty amino acid chain,” said Mohammad Heiranian, a Ph.D. candidate leading the research. “Our team is using nanopore-sequencing technology for protein detection to help determine single point mutations which can cause a variety of diseases. The goal is to identify the 20 essential amino acids with high precision and high resolution to aid in disease detection. Performing this research requires a fast, inexpensive way to identify the amino acids.”
“Our team uses supercomputers and machine learning (ML) to perform simulations in our amino acid research,” said Amir Taqieddin, another Ph.D. candidate. “Using supercomputers and ML provides a huge leap forward allowing our team to do experiments that are hard to do and run thousands of simulations, which would not be possible in our lab.”
The team used the Stampede2 supercomputer, one of the most powerful supercomputers in the U.S. for open science research, located at the University of Texas at Austin's Texas Advanced Computing Center (TACC) to run 4,293 simulations studying amino acids for 65 microseconds of molecular dynamics simulation—four orders of magnitude larger than typical simulation time.
“Due to the large amount of data and computation required, this work would take approximately 100 to 200 years of processing on a laptop or takes 50 years on a cluster computer,” said Taqieddin. “Our team was able perform over 4,000 amino acid simulations on Stampede2 in slightly over a month of computation time.”
Discovery of Defective Amino Acids with Nanopore-Sequencing
The team uses supercomputers running nanopore-sequencing technology for parallel analysis of thousands of protein pores with the ability to read a chain of DNA thousands of times. Biological nanopore sequencing uses transmembrane proteins, called porins, that cross a cellular membrane and act as a pore, through which molecules can disperse. The pores contain size dependent porous surfaces — with nanometer scale "holes" distributed across the membranes.
The nanopore has tiny holes and most materials used in nanopore sequencing are too thick, meaning that they span multiple amino acid chains, according to the scientific literature. This causes issues because the signal returned from simulation is from multiple amino acids rather than a single amino acid. An analogy in the real world might be a slot where a single soccer ball should fit. If the slot is too wide, then perhaps ten soccer balls would fall into the slot and the results of testing would be inaccurate for a single ball
The team used a nanoporous single-layer molybdenum disulfide (MoS2) which is a two-dimensional (2D) material in their research.
“The significance of MoS2 is that it is thin, only covering three atoms,” said Heiranian. “We can accurately identify the signal from a single amino acid to determine the properties of proteins. If simulations show the result of a faulty amino acid, then we know it is from a single, specific amino acid rather than multiple amino acids.”
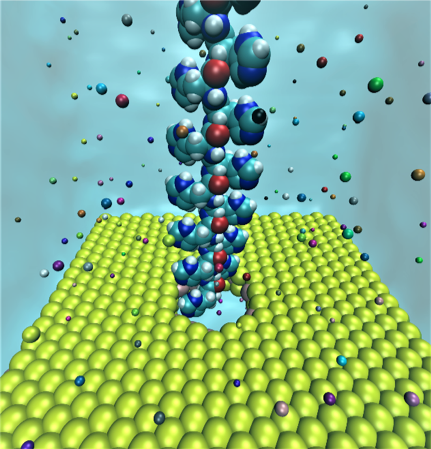
Figure 1. Simulation set up for the polypeptide chain with 16 units, MoS2 nanopore, and ions. Courtesy of University of Illinois, at Urbana-Champaign.
Supercomputers and Software used in the Research
The team used open source Nanoscale Molecular Dynamics (NAMD) software in their research. NAMD is noted for its parallel efficiency and is often used to simulate large systems containing millions of atoms. In addition, they used Intel MPI in their research which provided additional parallelization capabilities.
The TACC Stampede2 supercomputer used for the simulations is an 18-petaflop system containing 4,200 Intel Xeon Phi nodes, and it uses Intel Xeon Scalable processors and Intel Omni-Path Architecture.
“The scaling on Stampede2 was near ideal allowing us to complete our extensive simulations,” explained Heiranian.
Results of the Research
The nanopore research included 4,000 data points of the ionic current and resident time. Because of the volume of data, it was impossible to plot the whole domain for the different types of amino acids without doing millions of simulations. Using the Random Forest ML algorithm, they characterized the ionic current and residence time associated with the 20 standard amino acids by translocating them through a single-layer MoS2 nanopore using extensive simulations. Supervised and unsupervised machine learning and classification techniques were used to classify and detect signals with a high prediction accuracy of up to 99.6%.
Get the best insights in digital health directly to your inbox.
Related
Amazon and Pitt Health Data Alliance Strike Machine Learning Deal for Patient Care
Novartis Knew of Test Data Manipulation Before Drug Approval, FDA Says
Newsletter
Get the latest hospital leadership news and strategies with Chief Healthcare Executive, delivering expert insights on policy, innovation, and executive decision-making.