- Politics
- Diversity, equity and inclusion
- Financial Decision Making
- Telehealth
- Patient Experience
- Leadership
- Point of Care Tools
- Product Solutions
- Management
- Technology
- Healthcare Transformation
- Data + Technology
- Safer Hospitals
- Business
- Providers in Practice
- Mergers and Acquisitions
- AI & Data Analytics
- Cybersecurity
- Interoperability & EHRs
- Medical Devices
- Pop Health Tech
- Precision Medicine
- Virtual Care
- Health equity
3D Model Could Provide Better Understanding of Cancer's Spread
What might this means of data generation teach medicine about drugs?
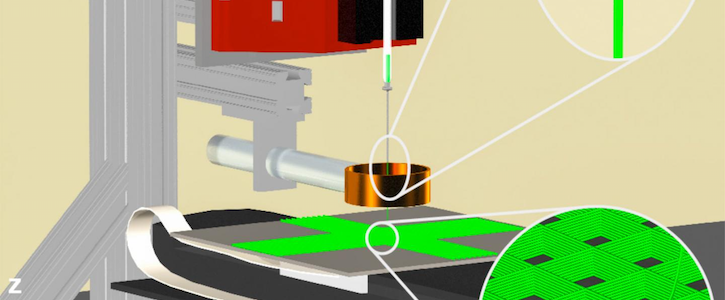
A mock-up of the 3D jet writer. Design has been cropped and resized. Courtesy of Purdue University/Luis Solorio.
Irrespective of the billions of dollars society spends on cancer research each year, scientists have yet to find a cure. In the United States alone, 1.7 million people are diagnosed with the disease, and about 600,000 die, each year.
The main reason that it is so hard to find effective treatments, researchers have said, is because the disease is so personal to an individual. Although many factors contribute to the development of cancer, there’s a lack of understanding about the underlying molecular mechanisms that drive it. Precision medicine, of course, has been gaining steam, but it has yet to take off across healthcare.
Researchers from Purdue University in Indiana hope to further the effort to make cancer treatment more personalized by identifying which drugs are particularly effective against a patient’s cancer type. The group, led by Luis Solorio, PhD, assistant professor of biomedical engineering, created a lifelike cancer environment out of polymer to better analyze drugs that might stop the disease in its tracks.
In the past, studies have used a 3D printer to re-create a controlled cancer environment, but the copies were not realistic enough for drug testing. The 3D jet writer developed at Purdue enables a cancer microenvironment by producing polymer tissues on an authentic scale with pore sizes large enough for cells to enter just as they would in a human body.
Right now, cancer treatment is not curative, so the long-term plan is for the technique to identify which drugs are particularly effective against a cancer, Solorio said.
“Clinicians have tons of different drugs at their disposal, but they don't really have the tools in place to be able to select which drugs might actually work, because a lot of those drugs are going to just make the patient feel better but not give them clinical benefit,” Solorio noted.
In the study, researchers used a patient’s fluids that already contained cancer cells—like those from water on the lungs, which are drained and usually discarded—to grow the cells. Solorio said the findings “are exciting,” as the cancer cells grew well and even outgrew some of the other cells in that space. When the researchers tried to take those same samples and grow them using standard tissue culture practices, they either died or stopped growing. “When we test them in this defined protein metrics, that never happened,” Solorio added.
In this case, the cells not only maintained the proliferative space but also the receptors that clinicians can use to determine which drugs a patient should receive.
Solorio hopes the finding will give clinicians a tool that will help guide their decision-making, which is particularly hard after a cancer has metastasized, and the cells are not as responsive to drugs as they were in the initial tumor.
“The clinician has only a limited amount of information to use when selecting the best drug or drug combination for the patient after the cancer has metastasized. So what we are hoping to do is provide a tool that can be used to inform the clinicians about which drugs may perform the best for a particular patient by screening thousands of compounds using the patients own cancer cells,” he said.
Researchers began working on the project at the Biointerfaces Institute at the University of Michigan and the continued it at Purdue.
The study was recently published in the journal Advanced Materials.
Related
Texas Team Becomes First in US to Perform AR Sinus Surgery
Stryker Reaches Deal to Distribute 3D Systems' Surgical Modeling Tech
